よむ、つかう、まなぶ。
【参考資料3】【英版R4.1.17】Nippon AMR One Health Report (NAOR) 2020 (57 ページ)
出典
| 公開元URL | https://www.mhlw.go.jp/stf/newpage_23261.html |
| 出典情報 | 国際的に脅威となる感染症対策関係閣僚会議 薬剤耐性ワンヘルス動向調査検討会(第9回 1/17)《厚生労働省》 |
ページ画像
ダウンロードした画像を利用する際は「出典情報」を明記してください。
低解像度画像をダウンロード
プレーンテキスト
資料テキストはコンピュータによる自動処理で生成されており、完全に資料と一致しない場合があります。
テキストをコピーしてご利用いただく際は資料と付け合わせてご確認ください。
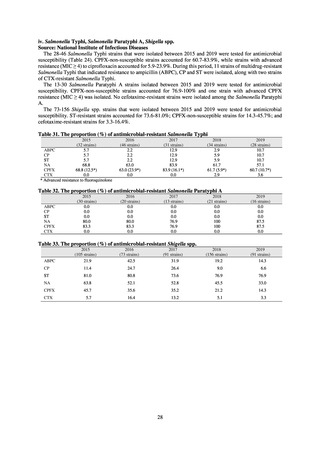
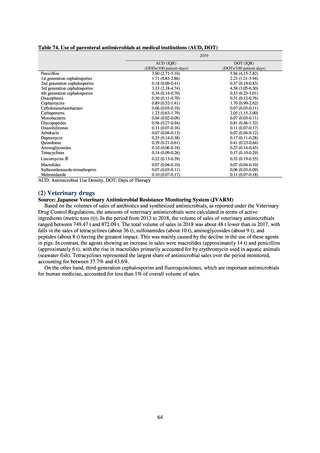
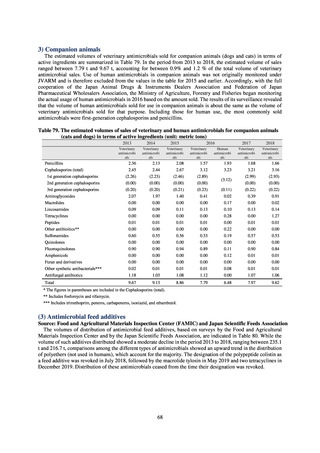
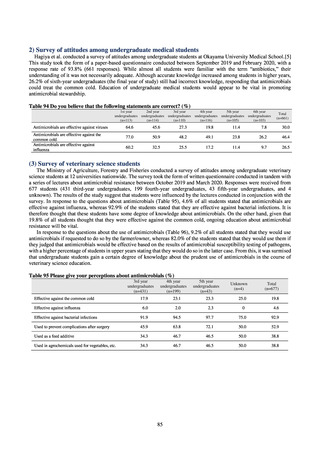
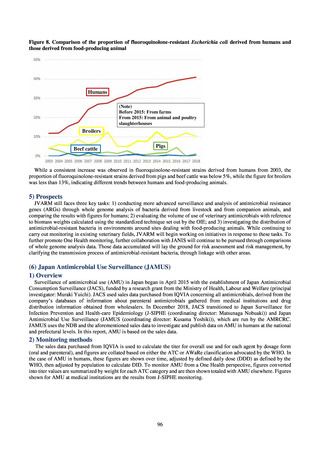
(3) Food
A 2019 research project to promote food safety, which was funded by a Ministry of Health, Labour and Welfare
research grant, found that the status of resistance among microbes isolated from food was as follows (FY2019
Health and Labour Sciences Research Grant General Report on the Research Project to Promote Food Safety:
Principal Investigator Haruo Watanabe). With the cooperation of 23 local public health institutes across Japan, the
research team used standardized methods to isolate strains of Salmonella contaminating food (mainly chicken)
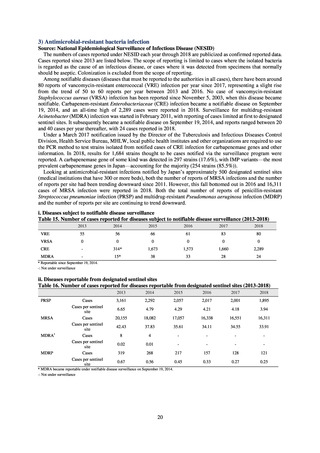
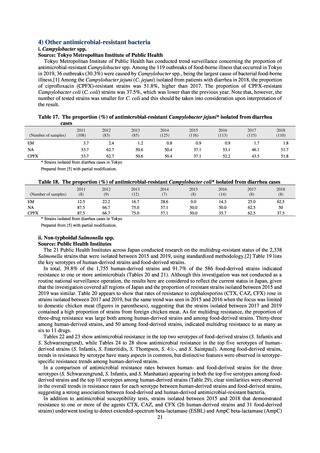
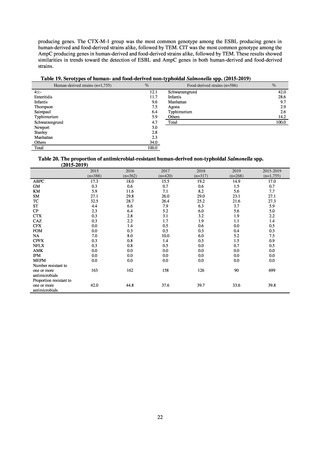
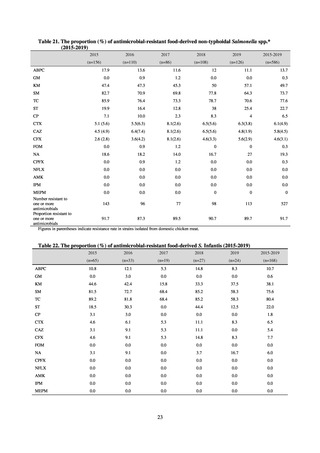
and to measure their antimicrobial resistance. These results are provided in 4) ii. Non-typhoidal Salmonella (local
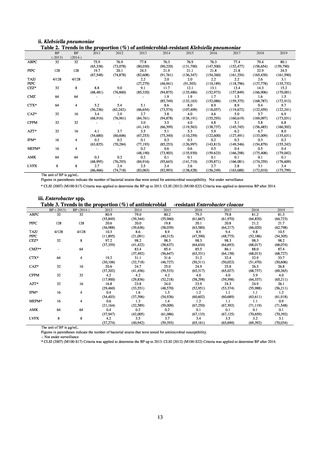
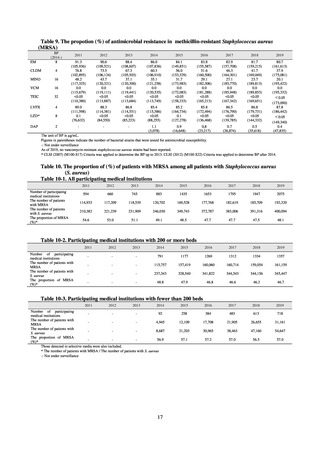
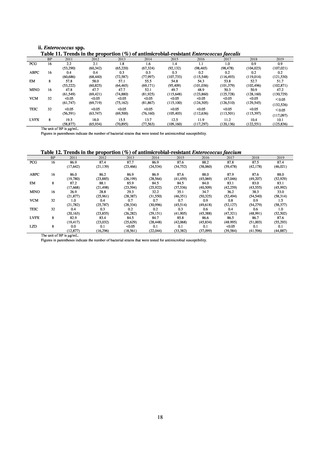
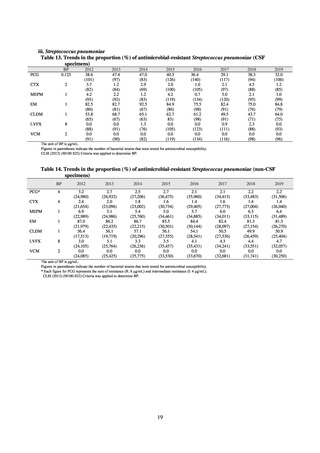
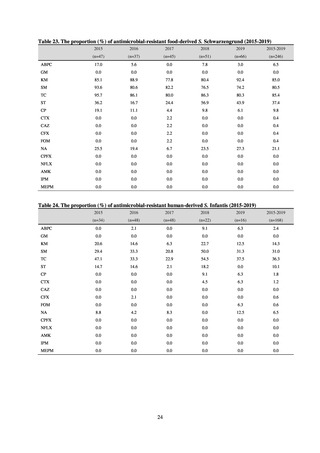
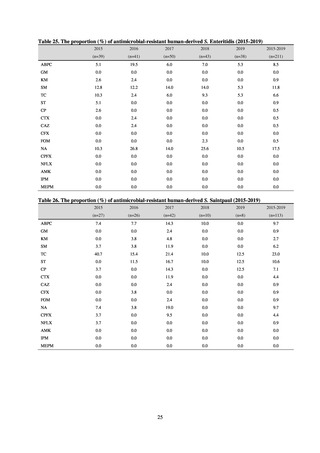
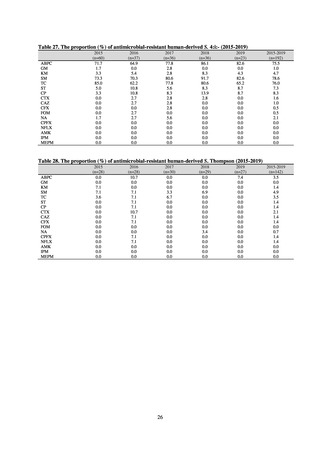
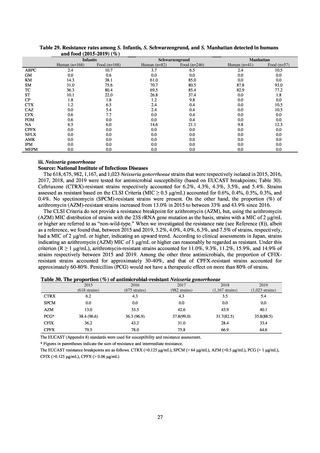
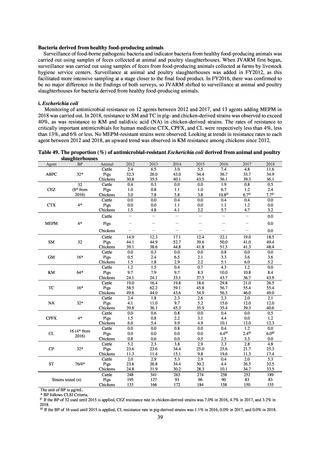
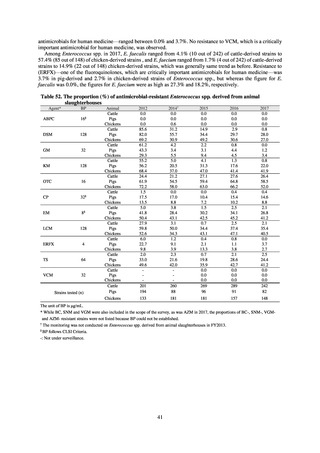
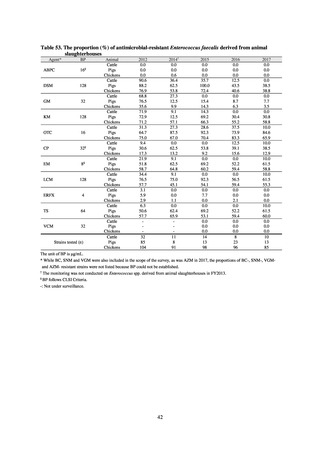
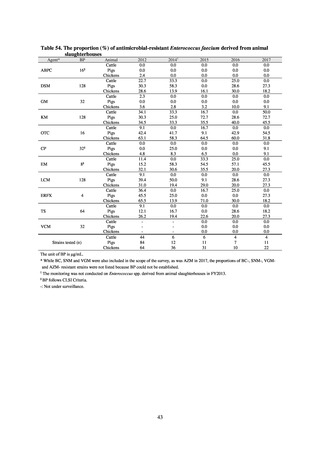
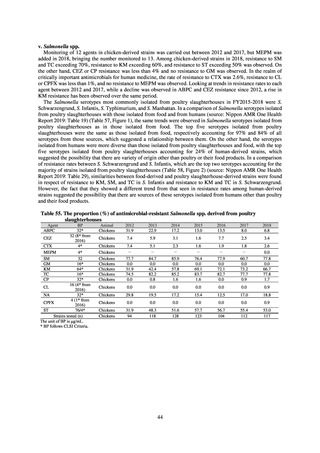
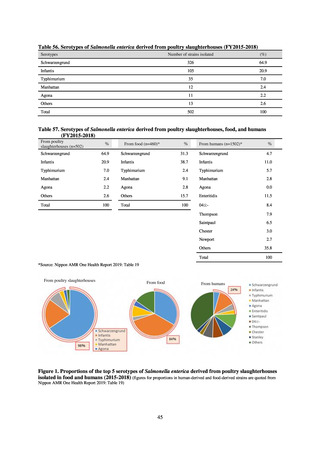
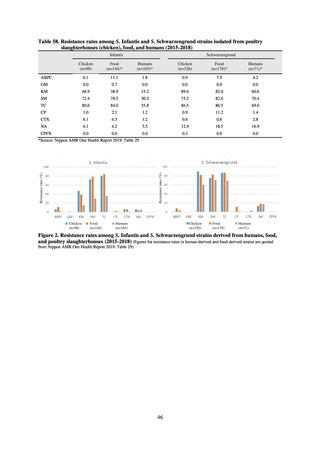
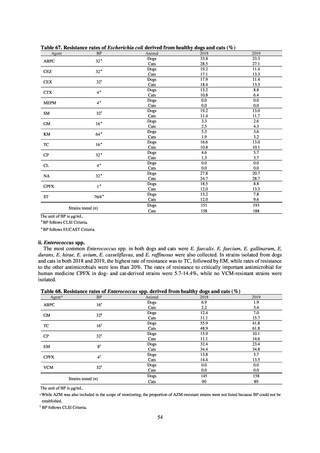
public health institutes) (see Tables 21, 22, 23, and 29). Comparisons of resistance rates among S. Infantis and S.
Schwarzengrund strains isolated from poultry slaughterhouses (chicken), food, and humans (2015-18) can be
found in Tables 57 and 58. In summary, the Salmonella serotypes and antimicrobial resistance patterns in samples
isolated from poultry slaughterhouses were observed to demonstrate the same tendencies as Salmonella in samples
isolated from food (primarily chicken). However, the Salmonella serotypes and antimicrobial resistance patterns
in samples isolated from humans (feces) were more diverse than the strains isolated from poultry slaughterhouses
and food, suggesting the possibility that there are diverse causes besides poultry and other food products (such as
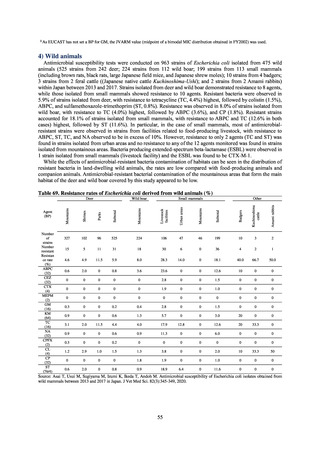
Salmonella derived from infections in turtles and other companion animals). In the case of Campylobacter, both
C. jejuni and C. coli showed strong similarities in terms of resistance trends between strains derived from human
patients and those derived from food, strongly suggesting a relationship between resistant bacteria derived from
food and those derived from human patients.
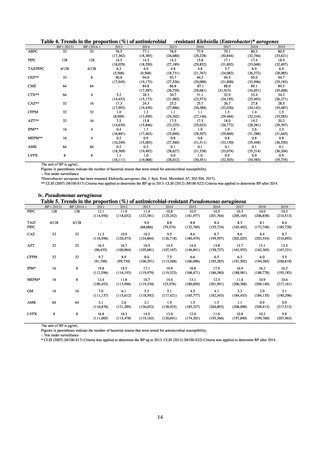
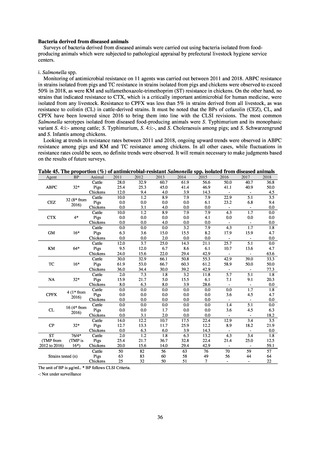
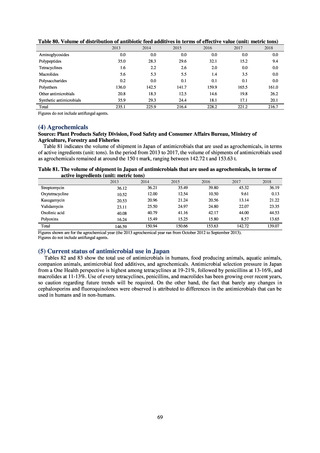
Antimicrobial susceptibility tests conducted on Escherichia coli strains isolated from commercially available
chicken meat found higher resistance to the following agents in strains isolated from domestic chicken meat: KM
(domestic 35.7%, imported 8.3%), TC (domestic 46.9%, imported 19.4%), ABPC (domestic 42.3%, imported
27.8%), CP (domestic 22.8%, imported 5.6%), ST (domestic 29%, imported 19.4%), and SM (domestic 37.3%,
imported 30.1%). On the other hand, resistance to NA (domestic 19.9%, imported 36.1%) and GM (domestic 5%,
imported 19.4%) was higher in imported chicken meat.
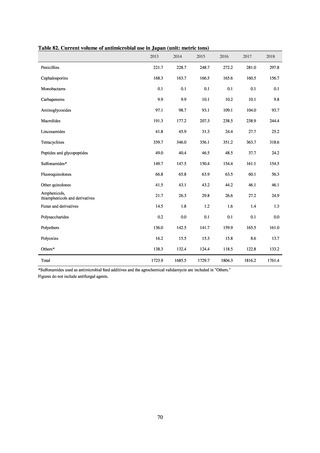
Testing of 311 Escherichia coli strains isolated from the feces of healthy individuals for susceptibility to 19
antimicrobial agents found that 39.2% of strains demonstrated resistance to at least 1 agent. The rate of resistance
to fluoroquinolones was 10%, while resistance to CTX was around 5%, following the same tendencies seen since
2015. No IPM- or MEPM-resistant strains were observed. Two strains were found to have the plasmid-mediated
colistin resistance gene (both were mcr-1 positive).
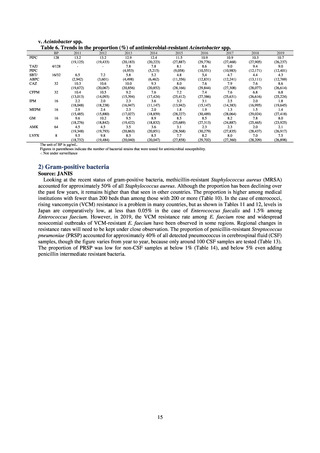
With the aim of detecting ESBL-producing Escherichia coli, 129 samples of domestic commercially available
chicken meat from FY2019 were selected for CTX resistance on a CTX-containing medium. CTX-resistant
Escherichia coli was found in 76.7% of chicken thigh meat samples and in 66.0% of chicken breast meat samples.
The highest number of CTX-resistant Escherichia coli bacteria per sample was approximately 3.0logCFU/g. While
it is not necessarily the case that Escherichia coli in chicken goes on to become established in the human intestine,
the fact that about 5% of Escherichia coli isolated from healthy individuals is CTX-resistant (almost all bacteria
had the ESBL gene) means one cannot deny the possibility that resistant bacteria or resistance genes are getting
into normal human flora via food. In future, it will be necessary to undertake a comparative analysis of resistant
bacteria from each source.
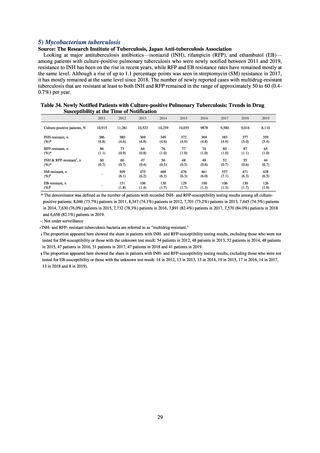
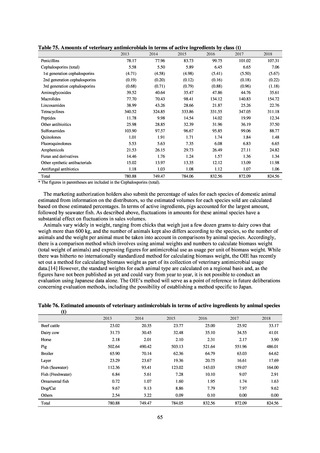
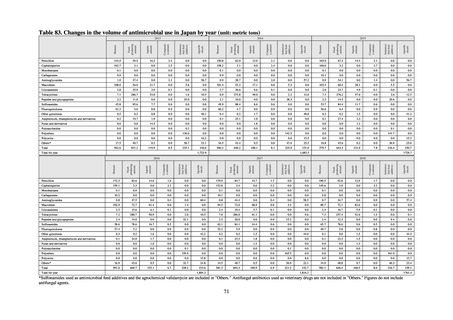
(4) Environment
In general, waste resulting from human activities is discharged into the environment (rivers or oceans) after
being treated at sewage treatment plants or other household wastewater treatment facilities until it meets effluent
standards. Attention to environmental AMR based on the One Health approach focuses on evaluating the risks
posed by antimicrobial-resistant bacteria (genes) by determining which antimicrobial-resistant bacteria (genes)
exist in environmental water discharged into the environment (rivers and oceans) after waste resulting from human
activities (rivers or oceans) is treated at sewage treatment plants or other household wastewater treatment facilities
until it meets effluent standards, and considering how those antimicrobial-resistant bacteria (genes) could circulate
into our daily lives and pose a risk to human health.
With few quantitative reports available at present concerning the extent to which antimicrobial-resistant bacteria
(AMR bacteria: ARB) and the antimicrobial-resistance genes (AMR genes: ARGs) that stem from them are
continuing to impose a burden after being excreted into the environment, a systematic nationwide survey is
regarded as essential. Accordingly, a research group funded by a Ministry of Health, Labour and Welfare research
grant has been formed for the purpose of conducting ongoing environmental AMR surveillance for the Japanese
government. Led by Hajime Kanamori, the research group is conducting a study entitled “Research to Establish
Methods of Surveying Antimicrobial-resistant Bacteria and Antimicrobials in the Environment” from 2018 to
2020.
56
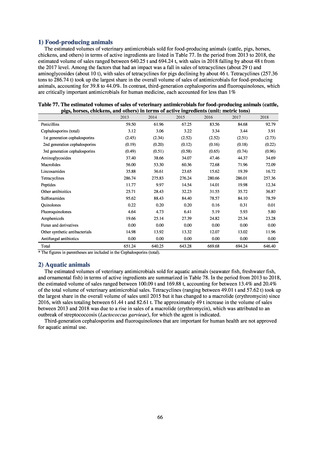
A 2019 research project to promote food safety, which was funded by a Ministry of Health, Labour and Welfare
research grant, found that the status of resistance among microbes isolated from food was as follows (FY2019
Health and Labour Sciences Research Grant General Report on the Research Project to Promote Food Safety:
Principal Investigator Haruo Watanabe). With the cooperation of 23 local public health institutes across Japan, the
research team used standardized methods to isolate strains of Salmonella contaminating food (mainly chicken)
and to measure their antimicrobial resistance. These results are provided in 4) ii. Non-typhoidal Salmonella (local
public health institutes) (see Tables 21, 22, 23, and 29). Comparisons of resistance rates among S. Infantis and S.
Schwarzengrund strains isolated from poultry slaughterhouses (chicken), food, and humans (2015-18) can be
found in Tables 57 and 58. In summary, the Salmonella serotypes and antimicrobial resistance patterns in samples
isolated from poultry slaughterhouses were observed to demonstrate the same tendencies as Salmonella in samples
isolated from food (primarily chicken). However, the Salmonella serotypes and antimicrobial resistance patterns
in samples isolated from humans (feces) were more diverse than the strains isolated from poultry slaughterhouses
and food, suggesting the possibility that there are diverse causes besides poultry and other food products (such as
Salmonella derived from infections in turtles and other companion animals). In the case of Campylobacter, both
C. jejuni and C. coli showed strong similarities in terms of resistance trends between strains derived from human
patients and those derived from food, strongly suggesting a relationship between resistant bacteria derived from
food and those derived from human patients.
Antimicrobial susceptibility tests conducted on Escherichia coli strains isolated from commercially available
chicken meat found higher resistance to the following agents in strains isolated from domestic chicken meat: KM
(domestic 35.7%, imported 8.3%), TC (domestic 46.9%, imported 19.4%), ABPC (domestic 42.3%, imported
27.8%), CP (domestic 22.8%, imported 5.6%), ST (domestic 29%, imported 19.4%), and SM (domestic 37.3%,
imported 30.1%). On the other hand, resistance to NA (domestic 19.9%, imported 36.1%) and GM (domestic 5%,
imported 19.4%) was higher in imported chicken meat.
Testing of 311 Escherichia coli strains isolated from the feces of healthy individuals for susceptibility to 19
antimicrobial agents found that 39.2% of strains demonstrated resistance to at least 1 agent. The rate of resistance
to fluoroquinolones was 10%, while resistance to CTX was around 5%, following the same tendencies seen since
2015. No IPM- or MEPM-resistant strains were observed. Two strains were found to have the plasmid-mediated
colistin resistance gene (both were mcr-1 positive).
With the aim of detecting ESBL-producing Escherichia coli, 129 samples of domestic commercially available
chicken meat from FY2019 were selected for CTX resistance on a CTX-containing medium. CTX-resistant
Escherichia coli was found in 76.7% of chicken thigh meat samples and in 66.0% of chicken breast meat samples.
The highest number of CTX-resistant Escherichia coli bacteria per sample was approximately 3.0logCFU/g. While
it is not necessarily the case that Escherichia coli in chicken goes on to become established in the human intestine,
the fact that about 5% of Escherichia coli isolated from healthy individuals is CTX-resistant (almost all bacteria
had the ESBL gene) means one cannot deny the possibility that resistant bacteria or resistance genes are getting
into normal human flora via food. In future, it will be necessary to undertake a comparative analysis of resistant
bacteria from each source.
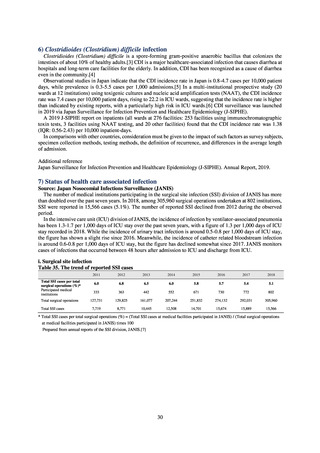
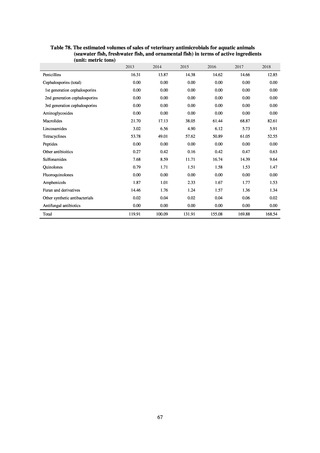
(4) Environment
In general, waste resulting from human activities is discharged into the environment (rivers or oceans) after
being treated at sewage treatment plants or other household wastewater treatment facilities until it meets effluent
standards. Attention to environmental AMR based on the One Health approach focuses on evaluating the risks
posed by antimicrobial-resistant bacteria (genes) by determining which antimicrobial-resistant bacteria (genes)
exist in environmental water discharged into the environment (rivers and oceans) after waste resulting from human
activities (rivers or oceans) is treated at sewage treatment plants or other household wastewater treatment facilities
until it meets effluent standards, and considering how those antimicrobial-resistant bacteria (genes) could circulate
into our daily lives and pose a risk to human health.
With few quantitative reports available at present concerning the extent to which antimicrobial-resistant bacteria
(AMR bacteria: ARB) and the antimicrobial-resistance genes (AMR genes: ARGs) that stem from them are
continuing to impose a burden after being excreted into the environment, a systematic nationwide survey is
regarded as essential. Accordingly, a research group funded by a Ministry of Health, Labour and Welfare research
grant has been formed for the purpose of conducting ongoing environmental AMR surveillance for the Japanese
government. Led by Hajime Kanamori, the research group is conducting a study entitled “Research to Establish
Methods of Surveying Antimicrobial-resistant Bacteria and Antimicrobials in the Environment” from 2018 to
2020.
56